Calculates the number of nodes, in each tree, that are common (i.e.: that have the same exact list of labels). The correlation is between 0 (actually, 2*(nnodes-1)/(2*nnodes), for two trees with the same list of labels - since the top node will always be identical for them). Where 1 means that every node in the one tree, has a node in the other tree with the exact same list of labels. Notice this measure is non-parameteric (it ignores the heights and relative position of the nodes).
cor_common_nodes(dend1, dend2, ...)Value
A correlation value between 0 to 1 (almost identical trees)
See also
Examples
set.seed(23235)
ss <- sample(1:150, 10)
hc1 <- iris[ss, -5] %>%
dist() %>%
hclust("com")
hc2 <- iris[ss, -5] %>%
dist() %>%
hclust("single")
dend1 <- as.dendrogram(hc1)
dend2 <- as.dendrogram(hc2)
cor_cophenetic(dend1, dend2)
#> [1] 0.4272001
cor_common_nodes(dend1, dend2)
#> [1] 0.8421053
tanglegram(dend1, dend2)
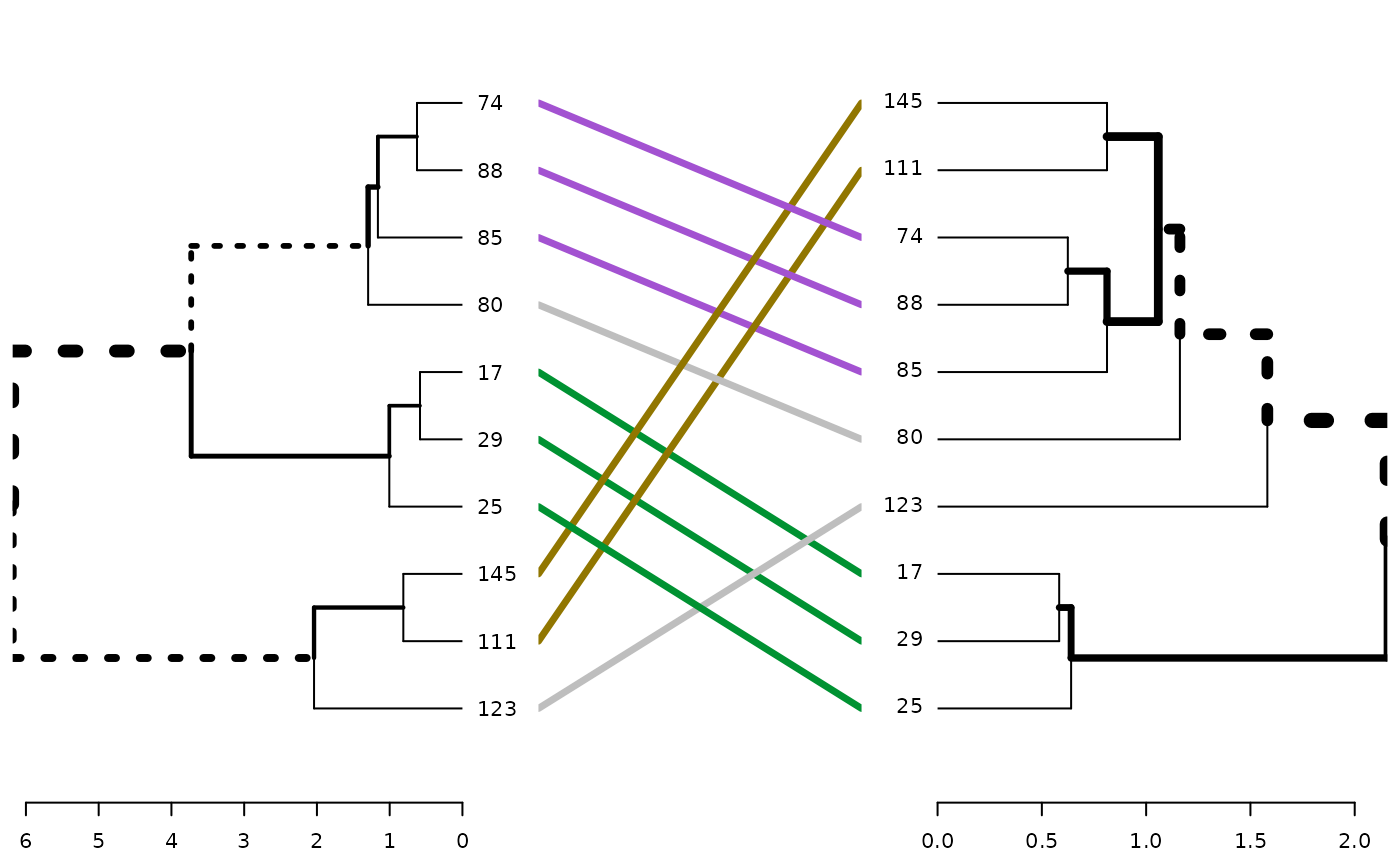 # we can see we have only two nodes which are different...
# we can see we have only two nodes which are different...