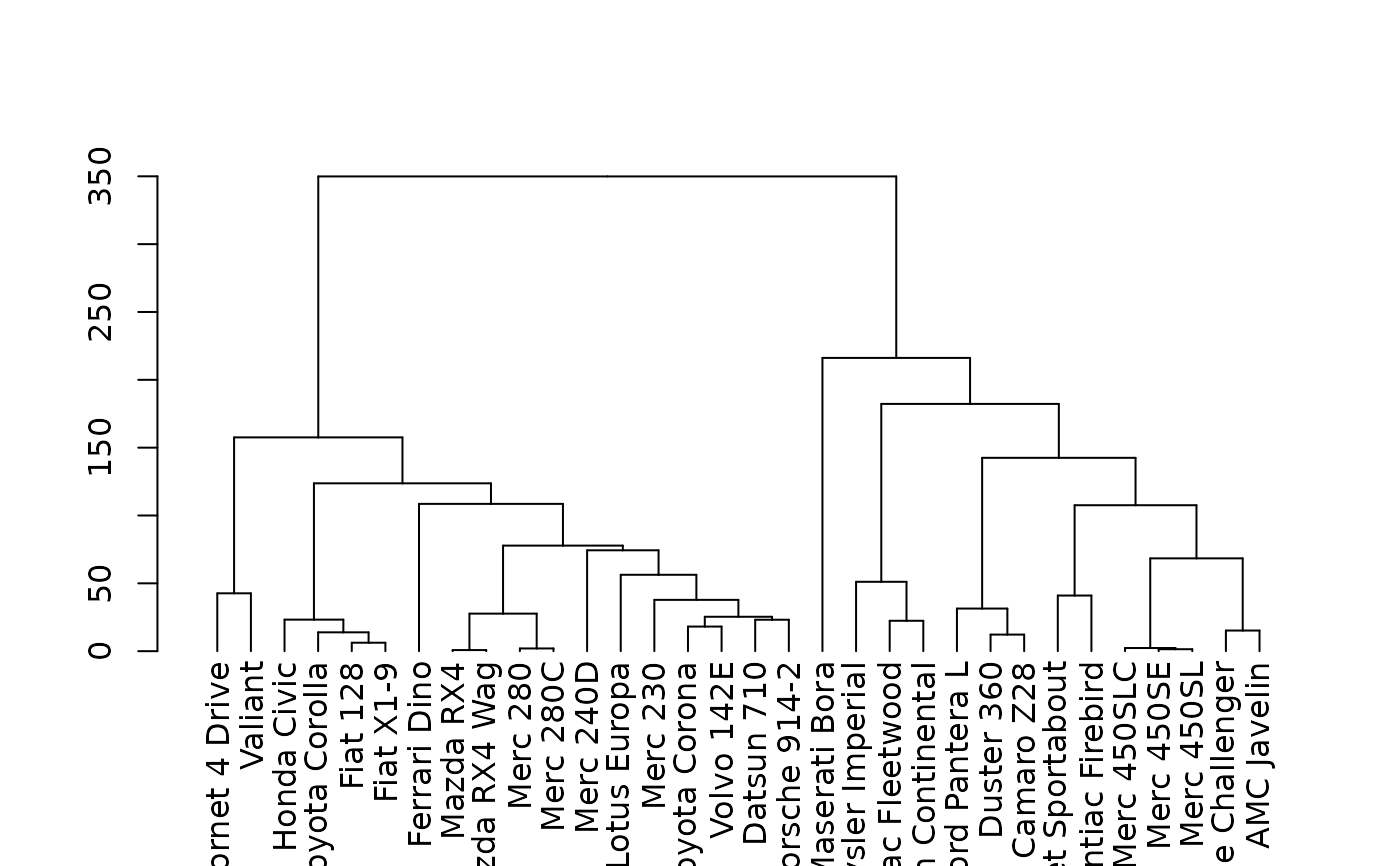
There are many options for choosing distance and linkage functions for hclust. This function goes through various combinations of the two and helps find the one that is most "similar" to the original distance matrix.
Arguments
- x
A matrix or a data.frame. Can also be a dist object.
- dist_methods
A vector of possible dist methods.
- hclust_methods
A vector of possible hclust methods.
- hclust_fun
By default hclust.
- optim_fun
A function that accepts a dend and a dist and returns how the two are in agreement. Default is cor_cophenetic.
- ...
options passed from find_dend to dend_expend.
Value
dend_expend: A list with three items. The first item is called "dends" and includes a dendlist with all the possible dendrogram combinations. The second is "dists" and includes a list with all the possible distance matrix combination. The third. "performance", is data.frame with three columns: dist_methods, hclust_methods, and optim. optim is calculated (by default) as the cophenetic correlation (see: cor_cophenetic) between the distance matrix and the cophenetic distance of the hclust object.
find_dend: A dendrogram which is "optimal" based on the output from dend_expend.
Examples
x <- datasets::mtcars
out <- dend_expend(x, dist_methods = c("euclidean", "manhattan"))
out$performance
#> dist_methods hclust_methods optim
#> 1 euclidean ward.D 0.7627152
#> 2 manhattan ward.D 0.7943439
#> 3 euclidean ward.D2 0.7778775
#> 4 manhattan ward.D2 0.8037024
#> 5 euclidean single 0.6834445
#> 6 manhattan single 0.7029393
#> 7 euclidean complete 0.8110543
#> 8 manhattan complete 0.8051400
#> 9 euclidean average 0.7935237
#> 10 manhattan average 0.8182836
#> 11 euclidean mcquitty 0.8114282
#> 12 manhattan mcquitty 0.8146632
#> 13 euclidean median 0.7888620
#> 14 manhattan median 0.8056585
#> 15 euclidean centroid 0.7917852
#> 16 manhattan centroid 0.8177614
dend_expend(dist(x))$performance
#> dist_methods hclust_methods optim
#> 1 unknown ward.D 0.7627152
#> 2 unknown ward.D2 0.7778775
#> 3 unknown single 0.6834445
#> 4 unknown complete 0.8110543
#> 5 unknown average 0.7935237
#> 6 unknown mcquitty 0.8114282
#> 7 unknown median 0.7888620
#> 8 unknown centroid 0.7917852
best_dend <- find_dend(x, dist_methods = c("euclidean", "manhattan"))
plot(best_dend)