Returns the set of all bipartitions from all edges, that is: a list with the labels for each of the nodes in the dendrogram.
partition_leaves(dend, ...)Source
A dendrogram implementation for partition.leaves from the distory package
Value
A list with the labels for each of the nodes in the dendrogram.
See also
distinct_edges, highlight_distinct_edges, dist.dendlist, tanglegram, partition.leaves
Examples
x <- 1:3 %>%
dist() %>%
hclust() %>%
as.dendrogram()
plot(x)
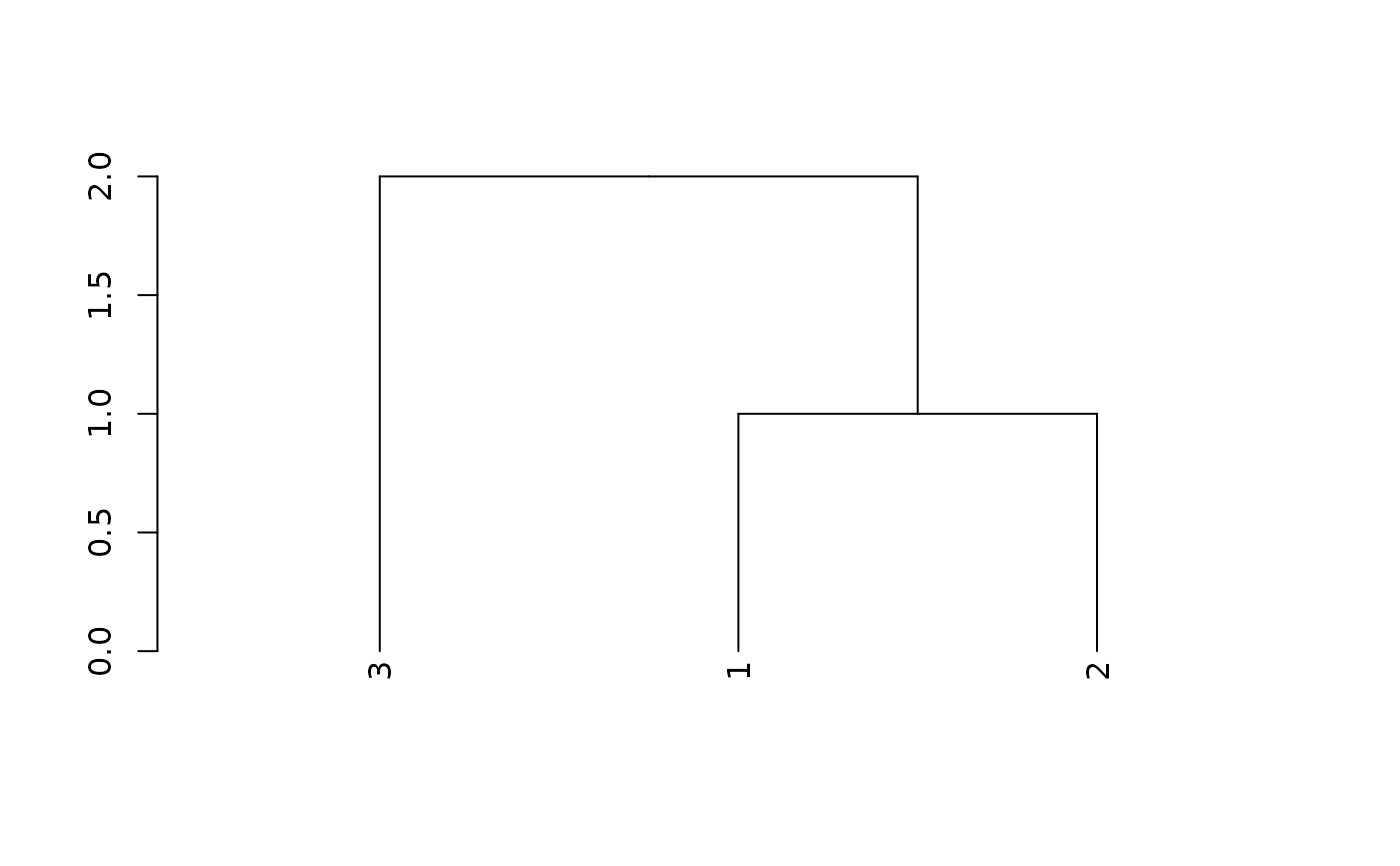 partition_leaves(x)
#> [[1]]
#> [1] 3 1 2
#>
#> [[2]]
#> [1] 3
#>
#> [[3]]
#> [1] 1 2
#>
#> [[4]]
#> [1] 1
#>
#> [[5]]
#> [1] 2
#>
if (FALSE) { # \dontrun{
set.seed(23235)
ss <- sample(1:150, 10)
dend1 <- iris[ss, -5] %>%
dist() %>%
hclust("com") %>%
as.dendrogram()
dend2 <- iris[ss, -5] %>%
dist() %>%
hclust("single") %>%
as.dendrogram()
partition_leaves(dend1)
partition_leaves(dend2)
} # }
partition_leaves(x)
#> [[1]]
#> [1] 3 1 2
#>
#> [[2]]
#> [1] 3
#>
#> [[3]]
#> [1] 1 2
#>
#> [[4]]
#> [1] 1
#>
#> [[5]]
#> [1] 2
#>
if (FALSE) { # \dontrun{
set.seed(23235)
ss <- sample(1:150, 10)
dend1 <- iris[ss, -5] %>%
dist() %>%
hclust("com") %>%
as.dendrogram()
dend2 <- iris[ss, -5] %>%
dist() %>%
hclust("single") %>%
as.dendrogram()
partition_leaves(dend1)
partition_leaves(dend2)
} # }